This documents the progress on my bioplastics project, which I conducted in the first half of 2019 at Chalmers Technical University in Gothenburg, Sweden. The goal here was to isolate interesting bacterial species from the environment, and investigate if they have the ability to produce specific chemical compounds – PHA (polyhydroxyalkanoates – that could be used in the production of biodegradable plastic. It’s a long road from finding a suitable bacterium to actually using it in an industrial-scale process, so here is a little glimpse into the first tiny steps on that journey.
Tuesday 15-01-2019
Today was all about preparing the plates that I will grow the bacteria on. Most bacteria like a mix of salt, proteins and sugar, and I prepared three different recipes of growth media to provide my future PHA producers with a bit of variety.
And look, it’s snowing outside! ❄︎❄︎❄︎❄︎

The nutritional broth contains agar, so after boiling it (which I want to do anyway to make everything sterile), it creates a jelly that the bacteria can grow on. I had to pour three litres of piping hot liquid into tiny petri dishes and let them cool down and solidify. I did that in the sterile work bench, otherwise I might get contamination (i.e. bacteria and fungi present in normal non-sterile environments) onto my plates.

Sunday 20-01-2019
It’s a misty, cold day and I go on a walk with my dog to take the first soil samples of what will be many more. The idea is to isolate bacteria from diverse environments (wet, dry, industrial, forest, etc) that contain a lot of rotting biomass (fields, forest floor, etc.) to get a diverse range of bacteria. I am in contact with forestry and agricultural companies that I will visit to take samples at their production and storage sites, but at the moment I am focusing on isolating samples from primarily fields and forests.




The samples are taken in sterile tubes and kept cool until I reach the lab, where I weigh a certain amount of each and suspend the soil in sterile water.


Finally, I make serial dilutions of each sample (from undiluted to 10^7 diluted) and spread it on the agar plates that I prepared previously. Then I incubate them all at a balmy 30°C and check again tomorrow.
Thursday 24-01-2019
We have growth!
After 24h of incubation, the bacterial colonies were still quite small, but 48h later I have some nice variety on the plates. Depending on the dilution, I got either a bacterial film covering the whole plate (not so useful…), or nicely distinct colonies of different colours, sizes and textures (much more useful!).
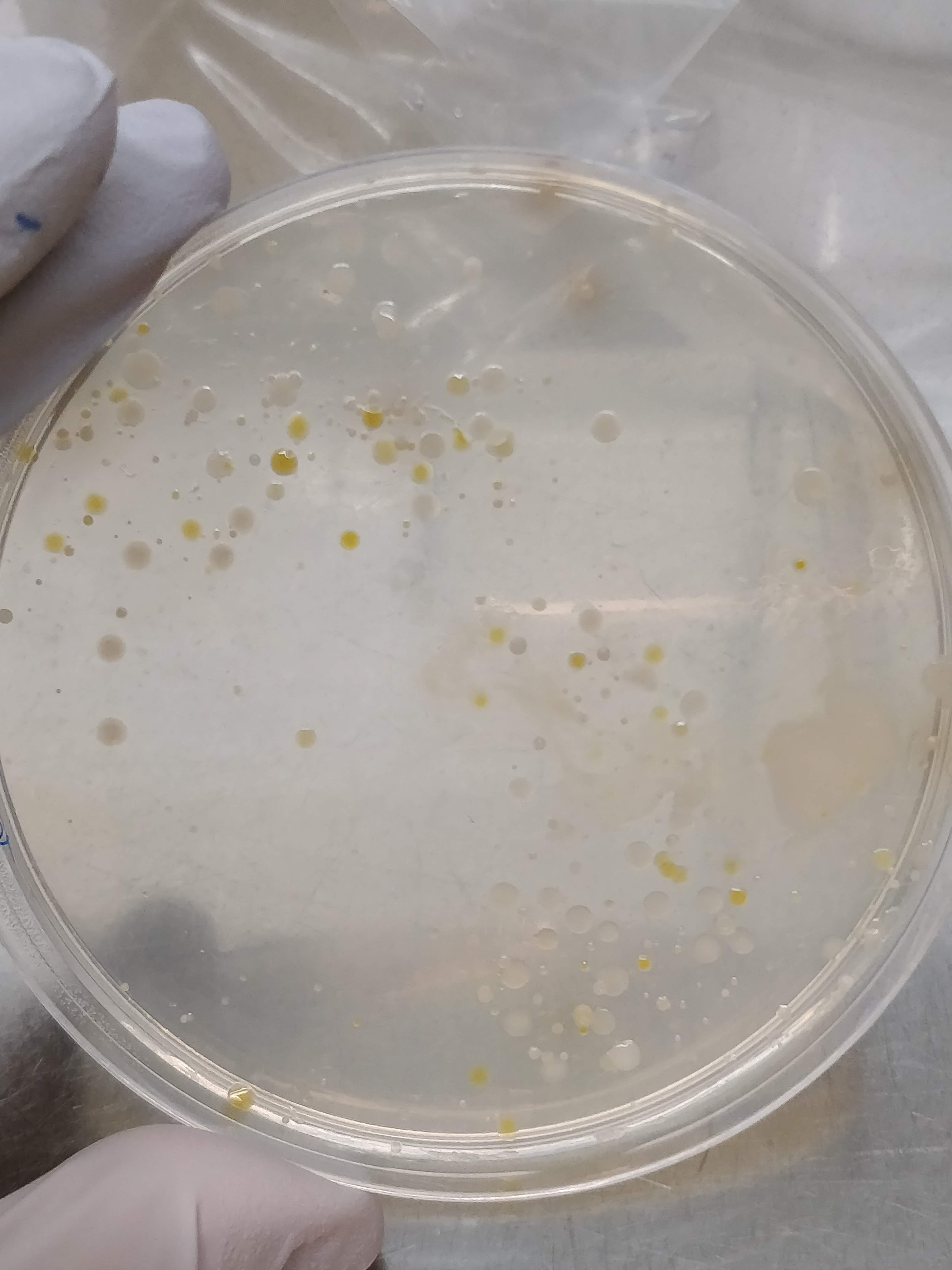
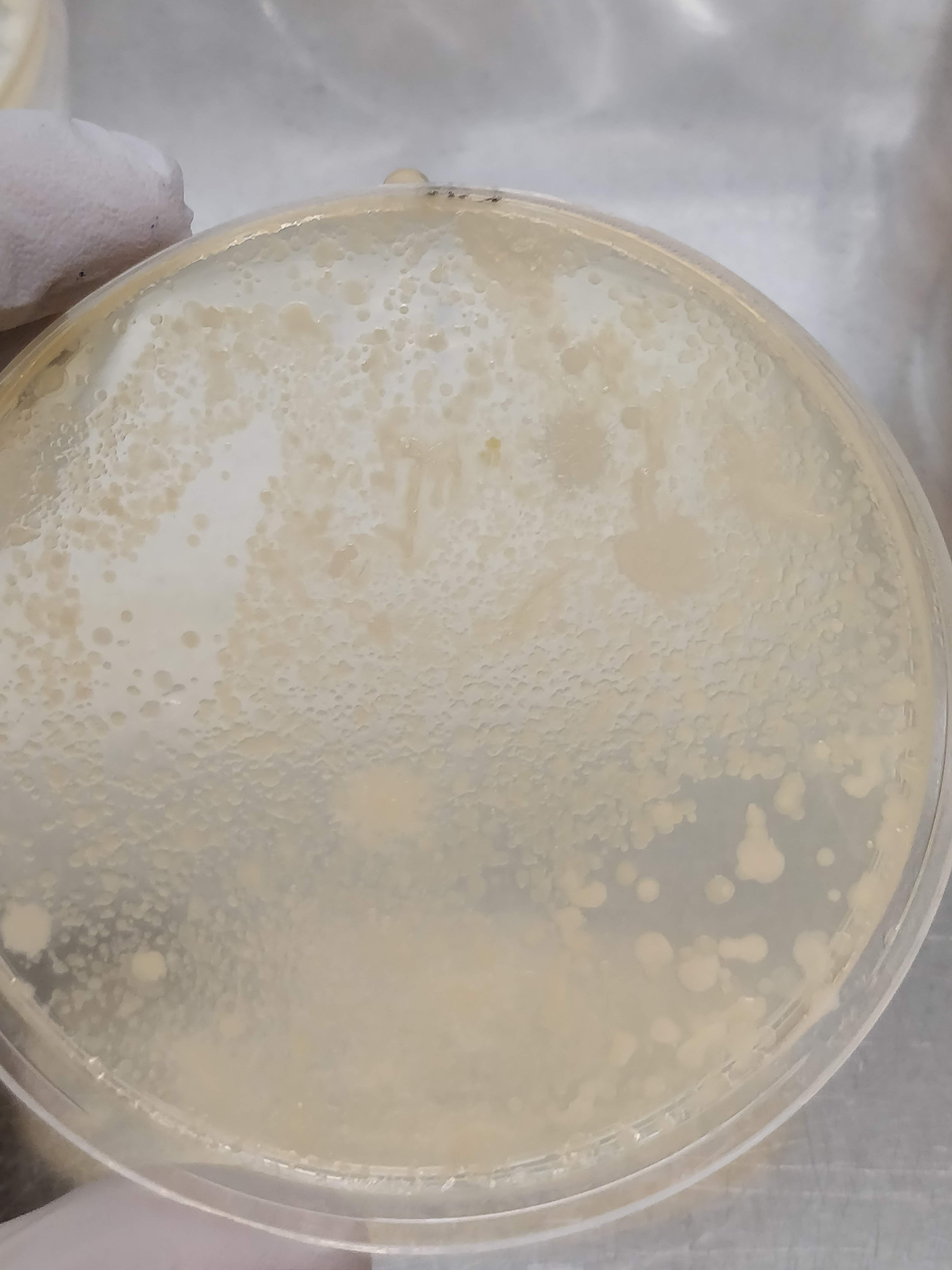
The next step is to create a master plate, which is a plate that collects many different bacterial colonies and serves as a kind of database for further experiments. To create this master plate, I dip a sterile pipette tip (or toothpick) into a colony and transfer it to a new plate. Then I do this about 200 times more.😴
Tuesday 29-01-2019
Here is one of the master plates! Isn’t it pretty? ☺️ I tried to pick as many different-looking colonies as possible, and as you can see on the master plate, some look really very different from the others. We have pink, light yellow, dark yellow, white, beige, dark purpel, etc. And also the way they grow looks very different.
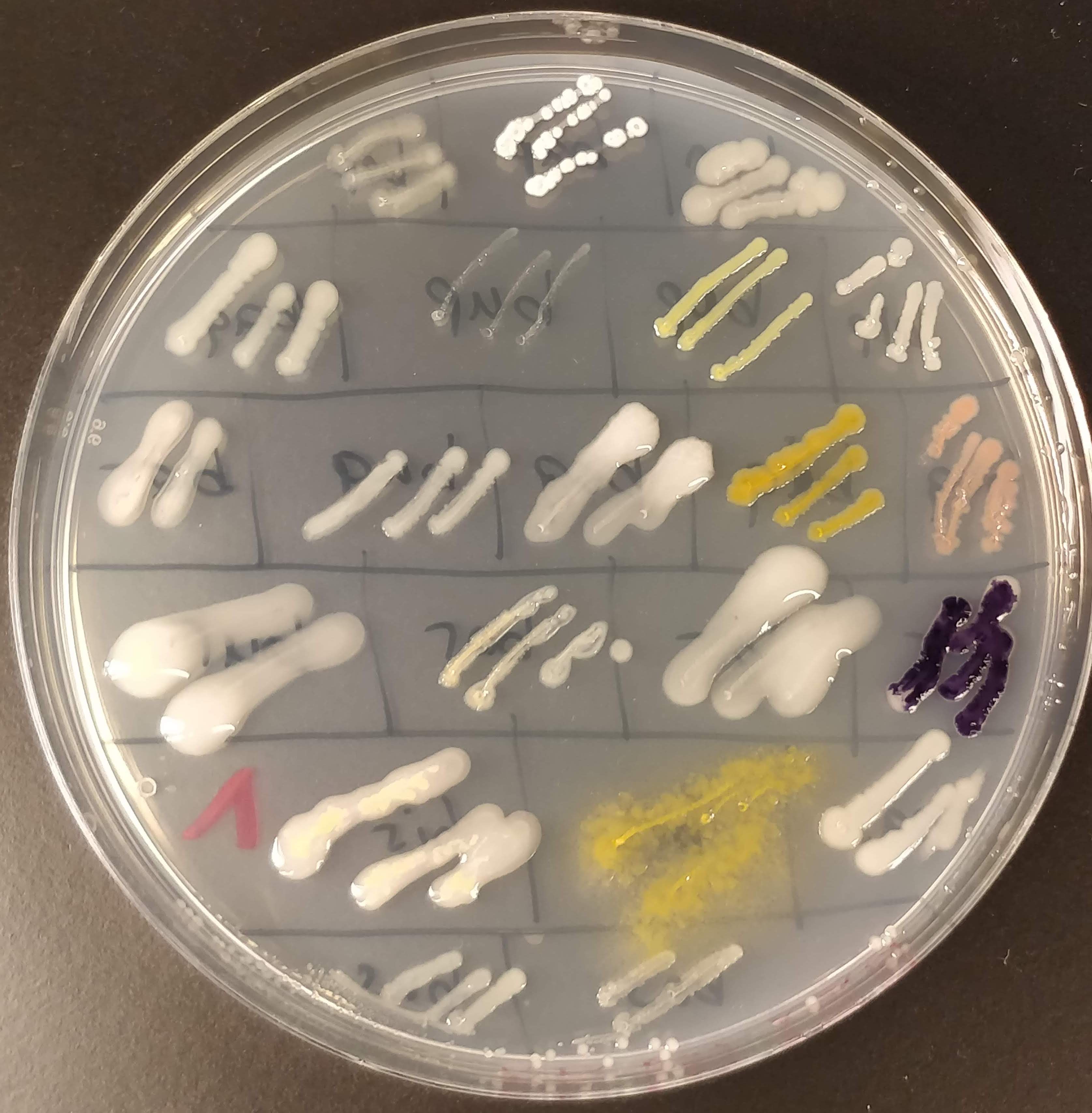
At this moment I have not much idea about what species the bacteria are and whether they produce anything interesting, like PHA. It’s unlikely that any of them are pathogenic, but to be sure, and also to avoid contamination from the outside, I only handle them inside the sterile work bench.
To distinguish the different bacterial samples, I have devised a naming convention that consists of a three letter combination of consonant-vowel-consonant. I find that easier than just numbering them. And it’s more fun if the best bacteria in the end are “cat” and “cow”. Or maybe “bob”.
To identify the bacterial species I would have to do a quite tedious procedure of growing each sample on different nutrients and at different conditions, look under the microscope, stain them with certain dyes, etc. Or I could just sequence their genome and compare the sequences to published databases. The first option is very time-intensive, the second option is quite costly. So for now I’m fine with not knowing exactly which species I have. That’s a task for later on, when I’ve narrowed the selection down to a few interesting specimen.
Thankfully I didn’t get much contamination on my plates. I added a low amount of cycloheximide to the medium to suppress fungal growth. I like fungi, but in this project I only want bacteria! The cycloheximide worked quite well, but on one plate I still got some filamentous growth. I will discard this plate, and transfer the unaffected colonies to a fresh plate.
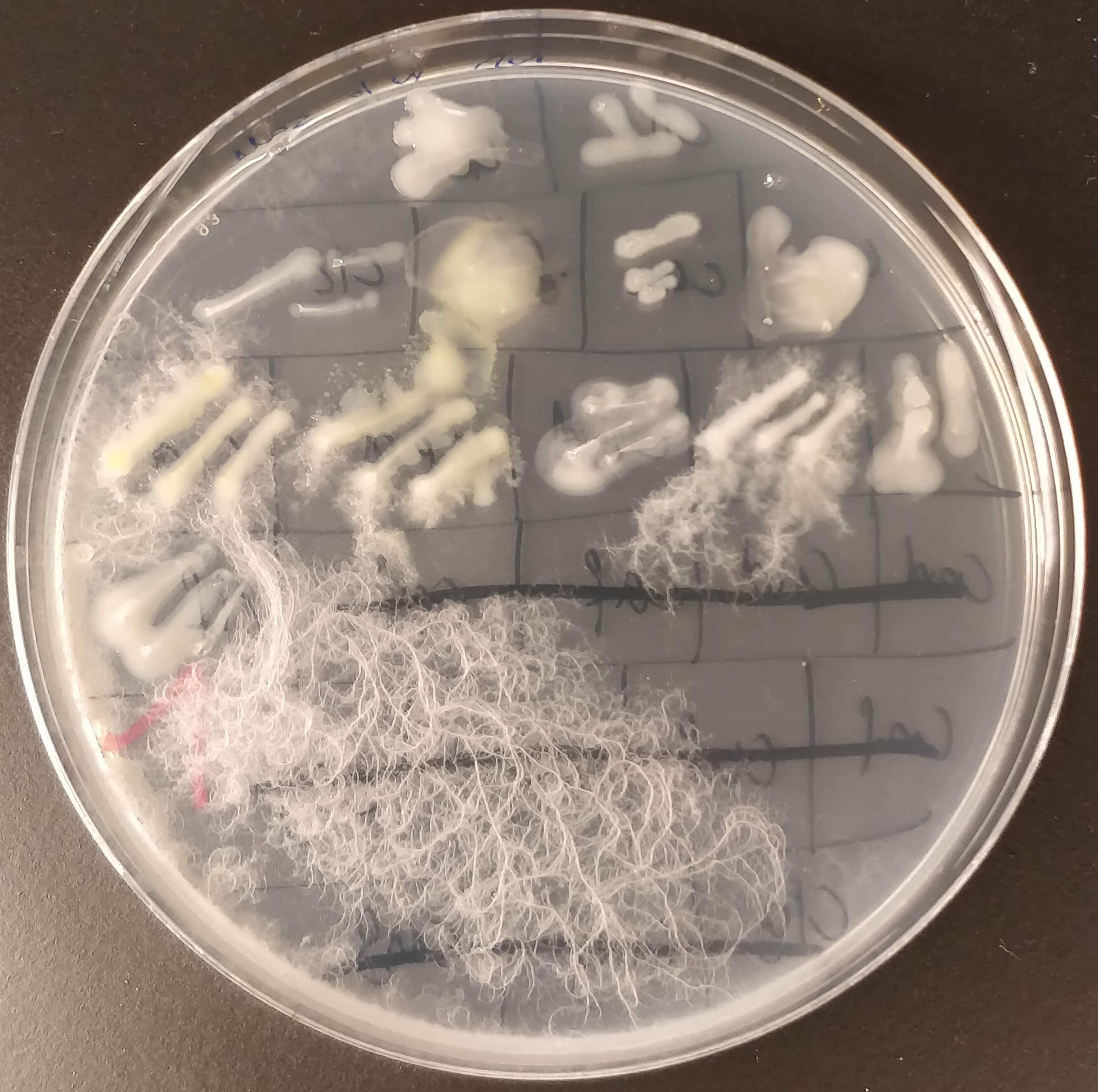
I do suspect that it’s bacteria that grow in this filamentous form, not fungi. There are bacteria that grow like this. And I’m sure they are super useful for some things, but I don’t really want them here. Sometimes, decision in science are also made out of pragmatic reasons. And in this project, I don’t want to deal with messy filamentous growth. So these candidates are out.
Wednesday 30-01-2019
Gothenburg is covered in snow, so for the time being I will not collect any new soil samples in the wild. Instead, I ordered some reference strains from the Culture Collection of Gothenburg University (CCGU) that I can use as positive controls. These reference strains are bacteria that are known to produce PHA, and research about them has been published. The bacteria arrived lyophilised in these little glass ampoules that you need to break in order to get the bacteria out.
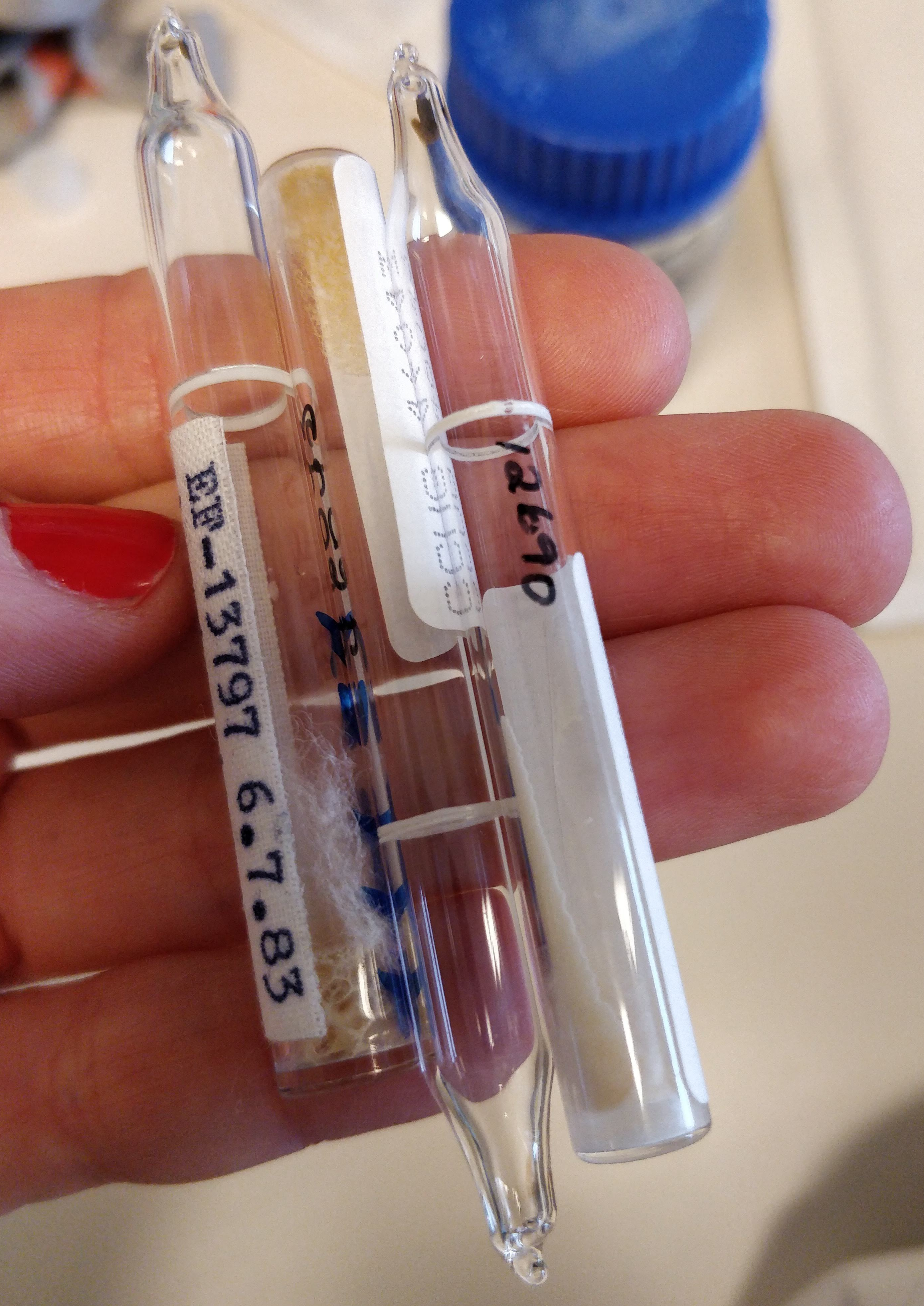
I had to make special growth medium for them, which contains beef extract and soybean meal. The resulting broth smelled like concentrated soy sauce and maggi. I hope the bacteria like it (I probably would!).
I also (again!) found these little creepy filamentous things growing on one of my plates. A couple more days of incubation, and I’m afraid they’ll escape the petri dish!

Tuesday 19-02-2019
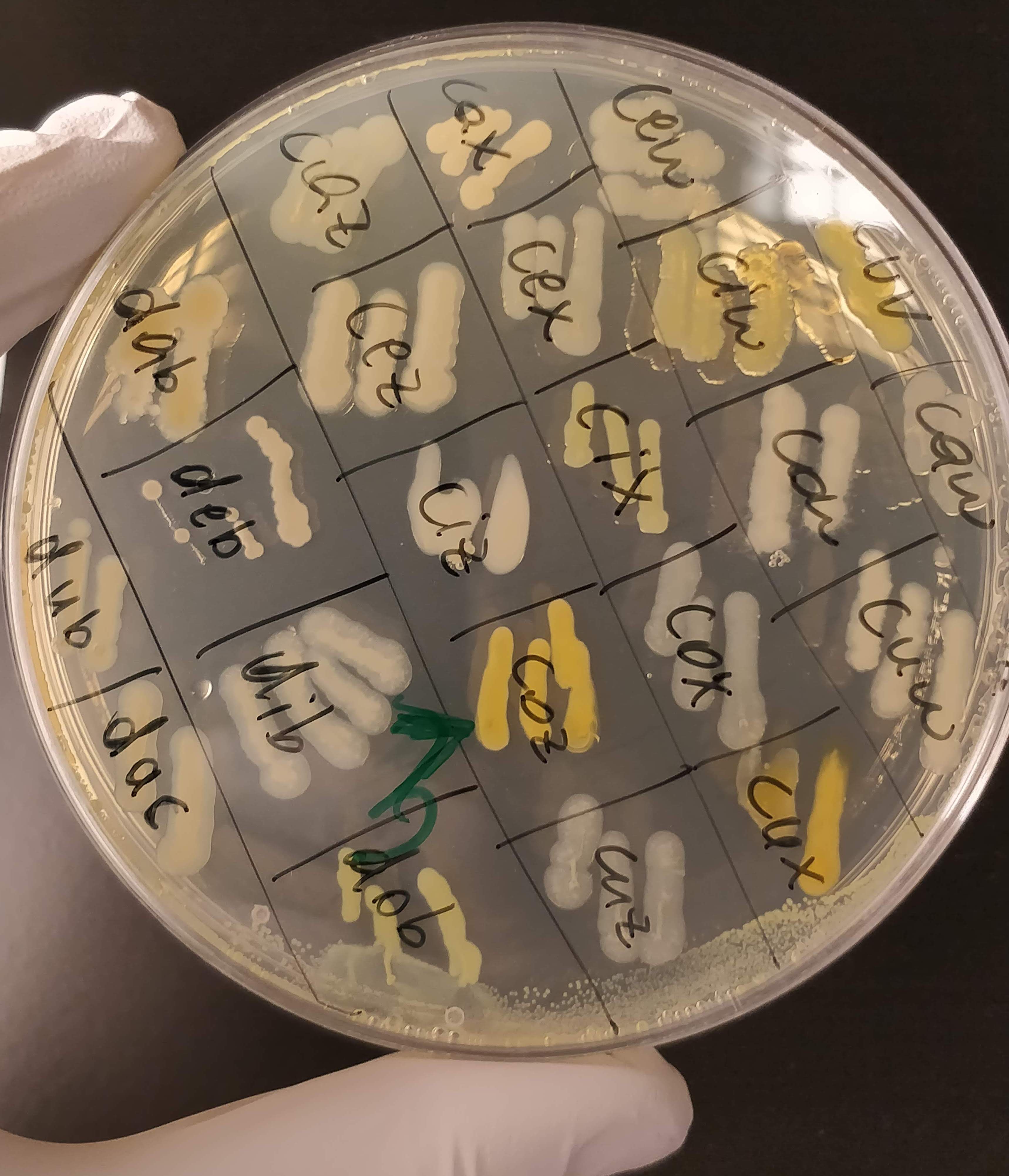
In the last couple of weeks I’ve been growing my bacteria (about 300 in total at the moment) on different substrates, to see what kind of sugars (glucose, sucrose, maltose, xylose, etc.) they like to eat. Most of them grow on several different kinds, which is a good sign! I also added a dye (Nile Red) into their growth medium that they take up during growth and binds to PHA inside the cell. The dye produces a strong orange fluorescence when it binds to PHA granules. And at the concentrations I use it, it should not inhibit cell growth at all. So the ones that glow under UV light are the strains that will continue to work with. I was a bit worried that a) none of them would grow and b) none of them would glow, but I got both in large amounts: GROW and GLOW!

Nile Red is a lipophilic dye that can also stain intracellular lipid droplets, so I am not totally sure yet if the glowing bacteria actually produce PHA. I can see a faint fluorescence for most strains that grew (e.g. 3caz or 3cez in the picture above), so it might be that that’s background fluorescence from lipids that are present in all bacteria, and that the ones that REALLY glow (like 3dax, 2cuj and 3cax) are the actual PHA-producers. I will have to run another kind of analysis to determine that.
